CHROMOSOMAL ABNORMALITIES IN SPONTANEOUS MISCARRIAGES IN A PUBLIC MATERNITY HOSPITAL IN THE CITY OF VITÓRIA, STATE OF ESPÍRITO SANTO, BRAZIL
Abstract
Objective: to verify the frequency of chromosomal abnormalities in miscarriage material from a public maternity hospital in the city of Vitória, state of Espírito Santo, Brazil. Method: A semi-direct culture of chorionic villi was performed. 70 samples were collected; 24 showed no chorionic villi and 46 samples passed the cytogenetic examination. Results: 31 samples did not achieve the cell growth stage and 15 samples have succeeded. Five normal karyotype and ten abnormal have been found, including trisomies, tetraploidy, monosomy X, triploidy and autosomal monosomy.
Conclusion: In this study the frequency of chromosomal abnormalities in spontaneous abortion material was high compared to similar studies in the literature.
Keywords: Spontaneous Abortion; Chromosomal Aberrations; Cytogenetics
Introduction
Abortion is a hemorrhagic syndrome that leads to conceptus death and/or expelling before it reaches its viability1. From the clinical standpoint, approximately 15% to 20% of pregnancies are spontaneously terminated.2,3 An abortion can be spontaneous, when there is no leading factor, or provoked, when there is a deliberate action for pregnancy termination. Considering the gestational age, it can also be classified into: early abortion, when it takes place up to the 12th week, and late, between the 12th and 22nd weeks, noting it usually happens within the first 12 weeks of pregnancy.4
Until the 1960s, the investigations carried out on fetus loss were limited to clinical and anatomic-pathological studies. The cytogenetics advent allowed the investigation of possible chromosomal anomalies in such cases4, which can be structural (isochromosome, translocation, inversion, deletion, insertion, duplication, ring chromosome) or numerical, and involve one or more autosomes, sexual chromosomes, or both.5 The numerical anomalies generally involve an extra chromosome, as in the Down Syndrome, for instance, in which occurs the trisomy of the chromosome 21 or the absence of a chromosome (monosomy), being the case of the Turner Syndrome, whose karyotype is 45,X.6 Besides the latter, there are other types of numerical alterations, such as triploidy, tetraploidy, tetrasomy and pentasomy.5
The chromosomal anomalies are present in about 50% of the spontaneous abortions of the first trimester.1-6 Due to the fact of many autosomal trisomies being incompatible with life, several embryos are spontaneously aborted. Many fetuses with autosomal trisomies may even survive, but their development is compromised. The Down Syndrome is the trisomy most compatible with life. Conversely, the monosomy of an autosome is always lethal, leading to a spontaneous abortion.6 Studies7-10 have spotted trisomies as the main causes for spontaneous abortions, followed by triploidies, monosomy X, tetraploidies, double trisomies and structural anomalies, not necessarily in this order.
The identification of a fetus loss cause helps to estimate the risks of recurrence in future generations, and by also promoting genetics counseling for the family. According to Horovitz et al,11 (2005), once excluded and proportionally rearranged the badly defined causes for child mortality, the perinatal ones held the first position in 1980 in Brazil, accounting for 38% of the deaths of infants, and the congenital anomalies occupying the fifth position, with 5% of the total. In 1990, this profile started to change, being observed a proportional reduction in the infectious and nutritional causes, leading the congenital anomalies to 8% and to take over the fourth cause. In the last year of evaluation (2000), the differences were substantially significant, with a great proportional reduction of deaths due to infectious and respiratory causes, now under 10%, and the congenital malformations holding the second position as a cause of the 13% of infant deaths. Such statistics on congenital defects and child mortality in Brazil can be seen as surprising, considering their magnitude and total lack of governmental policies with regards to their prevention and management.
In the face of a growing number of abortions of repetition and children born with congenital malformations, this study becomes relevant since it can contribute to the elaboration and/or improvement of public policies on this matter.
Objective
This paper’s objective is to verify the frequency of chromosomal anomalies present in spontaneous abortions in a public maternity hospital in the city of Vitória, state of Espírito Santo, Brazil.
Method
This study is characterized as prospective and was conducted with patients submitted to post-abortion curettage, of unknown reason, at the Pro-Matre Maternity Hospital, located in the municipality of Vitória, state of Espírito Santo, between April and October of 2007. Patients claiming to have provoked abortion have been ruled out of the study. After being made aware of all the necessary research information, if agreed, the patients signed the Informed Consent Form (ICF).
During curettage, ovular leftovers were collected in flasks containing saline solution, under aseptic conditions, separating the most prominent portion of amnion and placenta. The surgical center staff was also made aware of the necessary information for the most proper material collection. The labeled flasks were taken to the Cytogenetics Laboratory at the Federal University of Espírito Santo, by respecting the 48-hour deadline after sample collection.
The samples were submitted to a semi-direct culture of chorionic villus (24-48 hour culture), The villi present in the ovular leftovers were separated into sterile Petri plates. All procedures were conducted inside a biosafety cabinet (Bio Protector VECO®). After separated, villi were kept in previously labeled and prepared culture flasks as a means of culture (AmniomaxTM Gibco® INVITROGEN) by utilizing 4.2ml of the solution. The culture flasks were sealed and put into warm water bath at 37ºC/98.6ºF, for 24 hours. After that, the cultures were removed and treated with Colchicine (SigmaALDRICH®) and once again submitted to the warm water bath. After 45 minutes, the sample supernatant was collected with the aid of a Pasteur pipette and 5ml of a hypotonic solution (sodium citrate 0.075M) were added to the cultures, which would then be returned to the warm water bath for more 30 minutes at 37ºC/98.6ºF. For fixation, three baths were performed with [Methanol (Merck®) and acetic acid (Merck®)] in the 3:1 proportion, respectively, in a 10-minute interval. The fixing solution of the last bath was not discharged. The flasks with cultures were kept in a fridge for the preparation of slides in the following day. The slides (Precisium®) were previously washed with mild soap and kept in alcohol 92.8%. After drying and labeling with the patients’ data, they were put on a plate heated to 60ºC/140ºF. For each patient, an average of eight slides was prepared. The villi were then removed from the solution with a Pasteur pipette and put on a Petri plate in a scattered fashion and later addition of 2ml of a 60% acetic acid, allowing it to act for 2 minutes. After that, only the Petri plate material would be carefully dropped on each slide. The slides would only be removed from the heated plate after the complete drying process was complete. They were also submitted to the GTG banding, which consisted in a submersion for 1s in a 1:250 Trypsin solution (Gibco® INVITROGEN), and then washed in a phosphate buffer solution. Staining was performed by utilizing a 30% Giemsa stain, in which slides would be immersed for 7 minutes and then washed with distilled water.12
Cellular growth was assessed with the aid of an optical microscope. An average of fifteen to twenty metaphases was analyzed per case, which were manually drawn, and after a thorough assessment, the diagnosis would be issued. The absence of cellular growth was informed in the research results. The reports were compiled and mailed to the patients. Subjects who claimed to need further information on their results showed up at the Laboratory for a more detailed explanation of the diagnosis.
Results
70 samples of ovular leftovers were collected, being 24 out of those did not display any chorionic villi. Out of the 46 semi-direct cultures performed with chorionic villi samples, in 31 (67%) of them cellular growth was not observed. Cellular growth and subsequent karyotype recognition were possible in 33% (15/46) of the samples submitted to cytogenetics test.
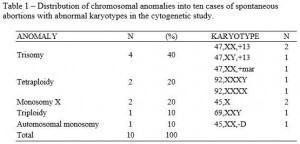
The cytogenetics study showed that 66% (10/15) of spontaneous abortion cases in the first trimester demonstrate chromosomal anomalies (Table 1). Three cases of chromosome 13 trisomy were found, compatible with the Patau Syndrome and one trisomy of a maker chromosome. Two cases of tetraploidy have also been found, besides two cases of monosomy X, compatible with the Turner Syndrome, and one case of triploidy. It was also found one case of monosomy of an autosomal chromosome of group D, karyotype 45,XX,-D. Out of the five cases found of normal karyotype, four were male (46,XX) and one female (46,XX).
Discussion
Since the 1970s it is technically possible to investigate chromosomal anomalies in abortion material.4 However, despite genetics’ great advancement, such analysis is performed in a few number of abortions in more developed centers.
In our study, 33% cellular growth success has been achieved. The literature suggests that up to 40% of cultures would not achieve cellular growth due to chorionic villi rapid degeneration.13 Nevertheless, taking into consideration that the present study was carried out with public hospital patients, it is possible that such percentage increase may have been caused by the long wait while the subjects had sought for medical assistance. Thus, the conception outcome was kept for a time longer than in the one where a sufficient amount of living cells could have been found, allowing us more cellular growth success and subsequent cytogenetics diagnosis.
Chromosomal anomalies are the most frequent known cause of a fetus death in our species. Its origin depends on several factors, including environmental and genetic reasons, besides an individual’s innate factor. Chromosomal anomalies can be classified into aneuploidies and euploidies. The former occur when the extra number of chromosomes is not an exact number of the haploid number, as a result of a proper non-disjunction or non-separation of one or more chromosomes during meiosis – as in the case of trisomy, during mitosis, when a cell divides itself into two daughter-cells, with a haploid number of chromosomes. The process of meiosis is to produce haploid cells from diploid cells. In monosomy X (45,X), X is present in the maternal chromosome in 70% to 80% of the cases; that is, in these cases, paternal X or Y are the ones lost during meiosis or in the first stages of embryogenesis.15 Euploidies are alterations that comprise the entire genome, originating cells that contain an exact multiple of 23 chromosomes in its nucleus, that is, an extra haploid group in a cell, besides the basic regular group of chromosomes. In turn, triploidies stem from maternal roots in approximately 80% of the cases, by the incorporation of a polar body into an oocyte. Less frequently, in about 20% of the cases, the origin is paternal, through dispermy, that is, the fertilization of an oocyte by two spermatozoa.15 Tetraploidies generally occur in the first miotic division (cleavage) of the zygote, in which chromosomes are regularly divided, but cytokinesis does not occur, being the result just one cell with 92 chromosomes.14 Rare cases have been described on the fertilization of one normal oocyte by three spermatozoa.16,17
In this study, the trisomies were the most common chromosomal anomalies and represented a 40% rate of the anomalies found, corroborating with several authors1-4,7-9,10,18, who have come to a result ranging from 30% to 50% in their studies. Among them, we found a karyotype with one marker chromosome, 47,XX,+mar., which is a structurally abnormal chromosome that cannot be identified by conventional banding techniques, such as the G band. From an unknown origin, it is present as an additional chromosome in the karyotype, either present or not in all cells. After additional investigations with molecular cytogenetic, for instance, the Fluorescent in situ hybridization (FISH) and array Comparative Genomic Hybridization (aCGH), it is now possible to find out the additional material origin.19,20 The FISH technique is based on the formation of a hybrid between DNA sequences or specific regions in cells or in chromosomes of cytological preparations and marked DNA probes. The formed hybrid can be directly observed through a microscope, after a marking with specific fluorochromes.19 The aCGH technique allows the investigation of gains and losses of DNA sequences of the entire genome.20
Other authors8,13 while studying cultures of chorionic villi have also found a rate ranging from 4% (16 in 380) to 4.2% (12 in 287) of tetraploidy. In our study the cases of monosomy of the X account for 20% (2 in 10) of the chromosomal anomalies found, these being similar to some studies7,19,21, whose rates are the ones of 21.4% (6 in 28); 23.7% (68 in 287) and 25% (75 in 305), respectively. Conversely, the study of Moraes et al. (2005) showed a significantly different rate, 4.2% (4 in 95). The studies’ closest results to our triploidy rate (10%, 1 in 10) are the ones of Alberman and Creasy (1977), with a rate of 13.2% (38 in 287) and the one by Perrone, Silva and Cintra (2006), with a rate of 15.5% (11 in 71). Regarding the case of monosomy of one autosomal chromosome of group D, there is no similar case in the literature. The chromosome belonging to this group are the acrocentric pairs of 13, 14 and 15. The formed embryo could have been developed by a monosomic gamete due to a non-chromosomal disjunction-involving chromosome D during maternal or paternal gametogenesis.22 Considering the material quality, it was not possible to identify to which pair it belonged, even when evaluating more than ten metaphases.
Although the cytogenetic technique is applied as the main conduct to screen and identify chromosomal anomalies present in most of the spontaneous abortions in the first trimester, an ideal practice is the combination of more state-of-the-art technologies, such as FISH, Multiplex Ligation-dependent Probe Amplification (MLPA) or aCGH, for a better understanding and more accurate diagnosis in these cases.23
Despite the sample analyzed being small, it was possible to observe that more than half of the cases of spontaneous abortions of the present study of cases have been caused by fetal chromosomal anomalies.
Several studies4,6,24,25 highlight the importance of the cytogenetic study of ovular leftovers and suggest that before gynecologists have couples undergo a series of additional and modern tests to investigate tentative loss causes, it would be of utmost importance to analyze the karyotype of such abortions.
Conclusion
In our study ten abnormal karyotypes have been found and the frequency of such chromosomal abnormalities determined. It was found among them trisomies, tetraploidies, triploidies, monosomies of the X and autosomal monosomy. The chromosomal anomalies have been found responsible for a high number of spontaneous abortions when compared to the literature. In these cases, the cytogenetic diagnosis assists in the understanding of the fetus loss and helps couples and physicians in a more complete and clarifying genetic counseling.
There has not been any conflict of interests in the present study.
References
1 Yakut S, Toru HS, Cetin Z, Özel D, Simsek M, Mendilcioğlu I, et al. Chromosome abnormalities identified in 457 spontaneous abortions and their histopathological findings. Turk Patoloji Derg. 2015;31(2):111-8.
2 Dhillon RK, Hillman SC, Morris RK, McMullan D, Williams D, Coomarasamy A, et al. Additional information from chromosomal microarray analysis (CMA) over conventional karyotyping when diagnosing chromosomal abnormalities in miscarriage: a systematic review and meta-analysis. BJOG. 2014;121(1):11-21.
3 Rabiega-Gmyrek D, Olejniczak T, Niepsuj-Biniás J, Guglas-Bochyńska B, Jachowski P, Latos-Bieleńska A, et al. Chromosomal aberrations – the cause of spontaneous abortions. Ginekol Pol. 2015;86(5):357-61.
4 Moraes AC, Moron AF, Hashimoto EM, Silva IDCG, Torloni MR, Souza MM, et al. Abordagem citogenética e molecular em material de abortos espontâneos. Rev Bras Ginecol Obstet. 2005;27(9):554-60.
5 Thompson MW, McInnes RR, Willard HF. Genética Médica. 5th ed. Rio de Janeiro: Guanabara Koogan, 1993. p.138-42.
6 Worton RG. Chromosome abnormalities: a major cause of birth defects, stillbirth and spontaneous abortion. Can Med Assoc J. 1977;117(8):849-51.
7 Uchida IA, Freeman VC, Gedeon M, Goldmaker J. Twinning rate in spontaneous abortions. Am J Hum Genet. 1983;35(5):987-93.
8 Alberman ED, Creasy MR. Frequency of chromosomal abnormalities in miscarriages and perinatal deaths. J Med Genet. 1977;14(5):313-5.
9 Procter SE, Watt JL, Gray ES. Cytogenetic analysis in 100 spontaneous abortions in North-East Scotland. Clin Genet. 1986;29(2):101-3.
10 Choi TY, Lee HM, Park WK, Jeong SY, Moon HS. Spontaneous abortion and recurrent miscarriage: A comparison of cytogenetic diagnosis in 250 cases. Obstet Gynecol Sci. 2014;57(6):518-25.
11 Horovitz DDG, Llerena Júnior JC, Mattos RA. Atenção aos defeitos congênitos no Brasil: panorama atual. Cad. Saúde Pública [online], Rio de Janeiro, BR,. 2005;21(4):1055-64.
12 GENOMA®. Laboratório de Genética do Espírito Santo. Cultura de vilo corial. Vitória, 2006.
13 Eiben B, Bartels I, Bähr-Porsch S, Borgmann S, Gatz G, Gellert G, et al. Cytogenetic analysis of 750 spontaneous abortions with the direct-preparation method of chorionic villi and its implications for studying genetic causes of pregnancy wastage. Am J Hum Genet. 1990;47(4):656-63.
14 Maluf SW, Félix TM, Schwartz IVD, Riegel M. Citogenética Humana. Porto Alegre: Artmed;2011. p.70-9.
15 Schinzel A. How human chromosome aberrations are formed. Atlas of Genetics and Cytogenetics in Oncology and Haematology. Poitier, France: Jean-Loup Huret, genetics DIM, University Hospital, 2007.
16 Sheppard DM, Fisher RA, Lawler SD, Povey S. Tetraploid conceptus with three paternal contributions. Hum Genet. 1982;62(4):371-4.
17 Surti U, Szulman AE, Wagner K, Leppert M, O’Brien SJ. Tetraploid partial hydatidiform moles: two cases with a triple paternal contribution and a 92,XXXY karyotype. Hum Genet. 1986;72(1):15-21.
18 Perrone DA, Silva DOM, Cintra TS. Frequência de Anomalias Cromossômicas em Abortos Espontâneos no Espírito Santo. Sogoes Revista. 2006;25:6.
19 Wei P, Li Y, Chen C, Zeng L, Qin S, Wang X, et al. Detection of chromosomal aneuploidies in spontaneous abortion samples by fluorescence in situ hybridization. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2015;32(2):229-32.
20 Li Y, Gong Y, Liu H, Song Y, He W, Wei J, et al. Detection for chromosomal aberrations in 43 fetuses with spontaneous abortion and stillbirth by array-based comparative genomic hybridization. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2015;32(3):348-52.
21 Kalousek DK. Anatomic and chromosome anomalies in specimens of early spontaneous abortions: seven-year experience. Birth Defects Orig Artic Ser. 1987; 23(1):153-68.
22 Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280-91.
23 Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A. 1986;83(9):2934-8.
24 Qumsiyeh MB. Chromosome abnormalities in the placenta and spontaneous abortions. J Matern Fetal Med.1998;7(4):210-2.
25 Hume H, Chasen ST. Trends in timing of prenatal diagnosis and abortion for fetal chromosomal abnormalities. Am J Obstet Gynecol. 2015;213(4):545.e1-4.
Acknowledgments
Our special gratitude to the patients that accepted to take part of this research, to the staff at the Pro-Matre Maternity Hospital for their collaboration and partnership, to the staff of the Service of Genetic Counseling at UFES and to all of whom, directly or indirectly, have collaborated with this study.
Attachments
Source: author. Note: translated.
Authors
Larissa Silva Zane¹*; Angela Maria Spagnol Perrone2; Iara Almeida Pinto3; Josivany Valério de Freitas Costa4; Marcela Souza Lima Paulo5; Agatha Cristhina Oliveira Faria6; Flavia Imbroisi Valle Errera7; Maria Regina Galvêas Oliveira Rebouças8;
1 Biologist – Master’s degree student at the Federal University of Espírito Santo (UFES). FAPES schorlarship.
2 Cytogeneticist Biologist – (Genoma Laboratory in Espírito Santo).
3 Pharmacist at the School of Sciences of Santa Casa de Misericórdia, EMESCAM, CNPq scholarship.
4 Biologist – PhD student – The Northeast Biotechnology Network – RENORBIO –Federal University of Espírito Santo – UFES.
5 PhD at the Federal University of Minas Gerais – UFMG – Professor of Medicine, Science and Technology at EMESCAM.
6 Pharmacist – PhD student in Sciences – Genetics (USP).
7 PhD in Sciences – Genetics (USP), Biologist – Professor of Genetics, Molecular Biology (EMESCAM).
8 Master in Sciences – Genetics (USP), Doctor Geneticist at HINSG and Professor of Genetics (UVV).